There are three examples below to help you manipulating the eddy files.
The examples can be downloaded here.
1: Allowing you the extraction of one eddy trajectory and store it in another file:
#!/usr/bin/env python
# -*- coding: utf-8 -*-
import xarray as xr
import numpy as np
from datetime import datetime
# Load dataset with xarray
with xr.open_dataset('eddy_trajectory_nrt_3.0exp_cyclonic_20180101_20190828.nc', decode_cf=False) as h: lon_min, lon_max, lat_min, lat_max = 1, 5, -28, -27 t = 25150 # Select a specific eddy with date and area, only some observations subset = h.sel(obs=(h.longitude > lon_min) & (h.longitude < lon_max) & (h.latitude > lat_min) & (h.latitude < lat_max) & (h.time == 25147)) # Extract full track subset = h.isel(obs=np.in1d(h.track, subset.track)) # Store selected data print(subset) subset.to_netcdf('output_eddy.nc')
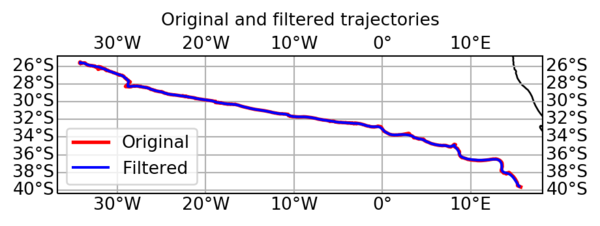
2: Allowing you plotting the selected eddy trajectory:
#!/usr/bin/env python # -*- coding: utf-8 -*- import xarray as xr from matplotlib import pyplot as plt # Load dataset with xarray ax = plt.subplot(111) with xr.open_dataset('output_eddy.nc') as h: N = 20 # plot contour every N ax.plot((h.speed_contour_longitude[::N].T + 180) % 360 - 180, h.speed_contour_latitude[::N].T, 'r') # plot path ax.plot((h.longitude + 180) % 360 - 180, h.latitude, 'b', label='eddy path') ax.set_aspect('equal') ax.legend() ax.grid() plt.show()
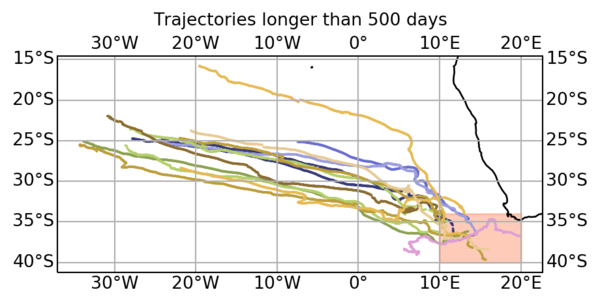
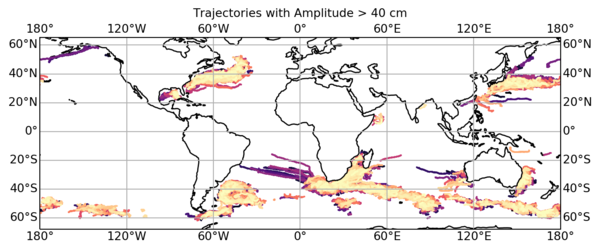
3: More advanced: Allowing you plotting a given eddy filtered track superimposed to its real track, and selecting all eddies on an area and having lifespan more than 500 days or amplitude >40cm: